News
[September 2024] I served as a judge in HopHacks. HopHack is a 36 hour annual Hackaton held at Johns Hopkins University. Teams create applications from scratch using available online APIs. The projects were first presented in a science fair format and were evaluated based on "usefulness", "polish", "creativity", and "technical difficulty". Out of over 60 projects, 10 were selected by the judges for a final presentation with live demos. Finally, the top 3 were selected for the awards. More information is available here and here.

[April 2024] I presented Movi, an efficient full-text index for pangenomes in Recomb-seq 2024. For more information look here and here. Find my slides here.

[February 2024] We released a new version of the Movi which exhibits even faster queries by implementing a latency hiding approach. We provide the new results in the new version of the preprint. Find it on BioRxv

[December 2023] I presented our latest work about using move structure for adaptive sampling of nanopore long reads in Genome Informatics 2023. Find my slides here.

[November 2023] We posted a pre-print about implementing move data stucture for cache-efficient string matching. Find it on BioRxv

[Fall 2023] I instructed a HEART course for incoming undergraduate students. The course was about an introduction to common exact and approximate string matching algorithms that are widely used for searching sequencing data. Find my slides here.

[August 2023] We posted a pre-print about classification of nanopore signal with a compressed pangenome index. Find it on BioRxv

[July 2023] Our paper about detecting isoform-level allelic imbalance by accounting for inferential uncertainty (SEESAW) was published in Genome Biology.

[May 2023] I presented our about indexing pangenomes with move structure in Workshop on Emerging Methods for Sequence Analysis at Penn State. Find my slides here.

[Fall 2022] I co-instructed (with Anna Liebhoff) a HEART course for incoming undergraduate students. The course was about an introduction to the application of software engineering in biomedical research.

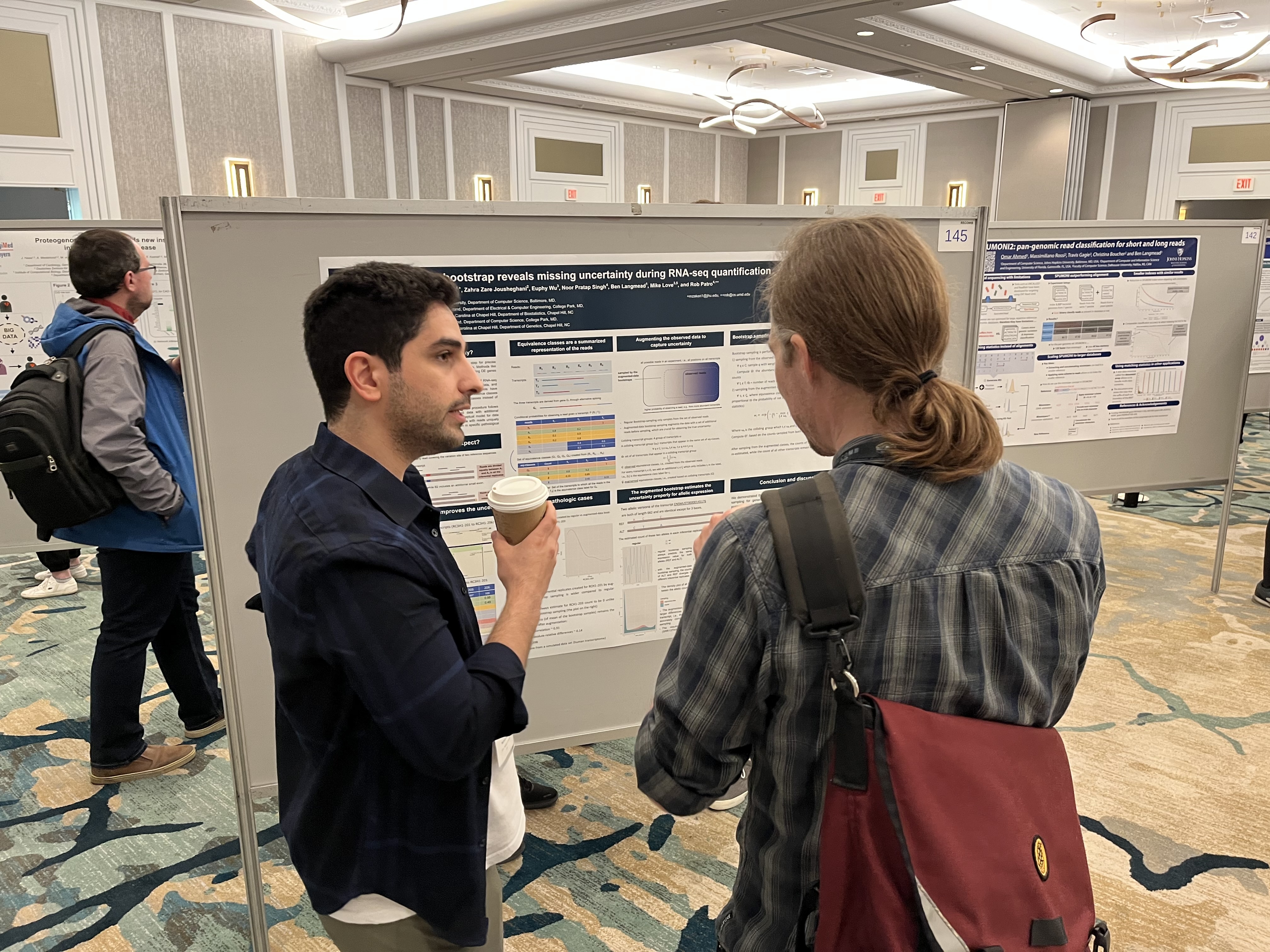
[May 2022] I presented a poster about computing RNA-seq quantification uncertainty with augmented bootstrap sampling in Recomb 2022. Find my poster here.
[March 2022] Our paper about a rapid, accurate, and memory-frugal tool for quantification of single-cell RNA-seq data (alevin-fry) was published in Nature Methods.

[February 2022] I joined Langmead-lab in Johns Hopkins University as a postdoctoral fellow.

[January 2022] Our paper about accurate quantification analysis of metahenomics and metatranscriptomics samples (AGAMEMNON) was published in Genome Biology.

[December 2021] I successfully completed my Ph.D. in Computer Science from the University of Maryland. During my Ph.D., I had the privilege of working in the Combine-Lab with Dr. Rob Patro as my PI. The title of my dissertation is "Optimizing the accuracy of lightweight methods for short read alignment and quantification". Find it here. You can also access my presentation slides here.
